Dealing with missing data in ANOVA models
June 25, 2018The analysis of variance, or ANOVA, is among the most popular methods for analyzing how an outcome variable differs between groups, for example, in observational studies or in experiments with different conditions.
But how do we conduct the ANOVA when there are missing data? In this post, I show how to deal with missing data in between- and within-subject designs using multiple imputation (MI) in R.
The ANOVA model
In the one-factorial ANOVA, the goal is to investigate whether two or more groups differ with respect to some outcome variable \(y\). The statistical model can be written as
\[ \begin{equation} \label{model} y_{ij} = \mu_j + e_{ij} \; , \end{equation} \]
where \(y_{ij}\) denotes the value of \(y\) for person \(i\) in group \(j\), and \(\mu_j\) is the mean in group \(j\). The (omnibus) null hypothesis of the ANOVA states that all groups have identical population means. For three groups, this would mean that
\[ \begin{equation} \mu_1 = \mu_2 = \mu_3 \; . \end{equation} \]
This hypothesis is tested by looking at whether the differences between groups are larger than what could be expected from the differences within groups. If this is the case, then we reject the null, and the group means are said to be “significantly” different from one another.
In the following, we will look at how this hypothesis can be tested when the outcome variable contains missing data. Let’s illustrate this with an example.
Example 1: between-subjects ANOVA
For this example, I simulated some data according to a between-subject design with three groups, \(n\) = 50 subjects per group, and a “medium” effect size of \(f\) = .25, which roughly corresponds to an \(R^2=6.8\%\) (Cohen, 1988).
You can download the data from this post if you want to reproduce the results (CSV, Rdata). Here are the first few rows.
| group | y | x | |
|---|---|---|---|
| 1 | A | -0.715 | 0.062 |
| 2 | B | 0.120 | 0.633 |
| 3 | C | 1.341 | 1.176 |
| 4 | A | NA | -0.792 |
| 5 | B | 0.468 | 0.243 |
The three variables mean the following:
group: the grouping variabley: the outcome variable (with 20.7% missing data)x: an additional covariate
In this example, cases with lower values in x had a higher chance of missing data in y.
Because x is also positively correlated with y, this means that smaller y values are missing more often than larger ones.
Listwise deletion
Lets see what happens if we run the ANOVA only with those cases that have y observed (i.e., listwise deletion). This is the standard setting on most statistical software.
In R, the ANOVA can be conducted with the lm() function as follows.
# fit the ANOVA model
fit0 <- lm(y ~ 1 + group, data = dat)
summary(fit0)#
# Call:
# lm(formula = y ~ 1 + group, data = dat)
#
# Residuals:
# Min 1Q Median 3Q Max
# -1.8196 -0.6237 -0.0064 0.5657 2.1808
#
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 0.0944 0.1452 0.65 0.52
# groupB 0.1720 0.1972 0.87 0.38
# groupC -0.1570 0.2082 -0.75 0.45
#
# Residual standard error: 0.895 on 116 degrees of freedom
# (31 observations deleted due to missingness)
# Multiple R-squared: 0.0229, Adjusted R-squared: 0.00609
# F-statistic: 1.36 on 2 and 116 DF, p-value: 0.26In this example, the \(F\)-test at the bottom of the output indicates that the group means are not significantly different from one another, \(F(\!\) 2, 116 \(\!)\) = 1.361 (\(p\) = 0.26).1 In addition, the effect size (\(R^2\) = 0.023) is quite a bit smaller than what was used to generate the data.
In fact, this result is a direct consequence of how the missing data were simulated. Fortunately, there are statistical methods that can account for the missing data and help us obtain more trustworthy results.
Multiple imputation
One of the most effective ways of dealing with missing data is multiple imputation (MI). Using MI, we can create multiple plausible replacements of the missing data, given what we have observed and a statistical model (the imputation model).
In the ANOVA, using MI has the additional benefit that it allows taking covariates into account that are relevant for the missing data but not for the analysis.
In this example, x is a direct cause of missing data in y. Therefore, we must take x into account when making inferences about y in the ANOVA.
Running MI consists of three steps. First, the missing data are imputed multiple times. Second, the imputed data sets are analyzed separately. Third, the parameter estimates and hypothesis tests are pooled to form a final set of estimates and inferences.
For this example, we will use the mice and mitml packages to conduct MI.
library(mice)
library(mitml)Specifying an imputation model is very simple here. With the following command, we generate 100 imputations for y on the basis of a regression model with both group and x as predictors and a normal error term.
# run MI
imp1 <- mice(data = dat, method = "norm", m = 100)The imputed data sets can then be saved as a list, containing 100 copies of the original data, in which the missing data have been replaced by different imputations.
# create a list of completed data sets
implist1 <- mids2mitml.list(imp1)Finally, we fit the ANOVA model to each of the imputed data sets and pool the results.
The analysis part is done with the with() command, which applies the same linear model, lm(), to each data set.
The pooling es then done with the testEstimates() function.
# fit the ANOVA model
fit1 <- with(implist1, lm(y ~ 1 + group))
# pool the parameter estimates
testEstimates(fit1)#
# Call:
#
# testEstimates(model = fit1)
#
# Final parameter estimates and inferences obtained from 100 imputed data sets.
#
# Estimate Std.Error t.value df P(>|t|) RIV FMI
# (Intercept) 0.025 0.145 0.171 2749.785 0.864 0.234 0.190
# groupB 0.204 0.200 1.022 4816.135 0.307 0.167 0.144
# groupC -0.328 0.204 -1.611 3274.908 0.107 0.210 0.174
#
# Unadjusted hypothesis test as appropriate in larger samples.Notice that we did not need to actually include x in the ANOVA.
Rather, it was enough to include x in the imputation model, after which the analyses proceeded as usual.
We now have estimated the regression coefficients in the ANOVA model (i.e., the differences between group means), but we have yet to decide whether the means are all equal or not. To this end, we use a pooled version of the \(F\)-test above, which consists of a comparison of the full model (the ANOVA model) with a reduced model that does not contain the coefficients we wish to test.2
In this case, we wish to test the coefficients pertaining to the differences between groups, so the reduced model does not contain group as a predictor.
# fit the reduced ANOVA model (without 'group')
fit1.reduced <- with(implist1, lm(y ~ 1))The full and the reduced model can then be compared with the pooled version of the \(F\)-test (i.e., the Wald test), which is known in the literature as \(D_1\).
# compare the two models with pooled Wald test
testModels(fit1, fit1.reduced, method = "D1")#
# Call:
#
# testModels(model = fit1, null.model = fit1.reduced, method = "D1")
#
# Model comparison calculated from 100 imputed data sets.
# Combination method: D1
#
# F.value df1 df2 P(>F) RIV
# 3.543 2 7588.022 0.029 0.188
#
# Unadjusted hypothesis test as appropriate in larger samples.In contrast with listwise deletion, the \(F\)-test under MI indicates that the groups are significantly different from one another.
This is because MI makes use of all the observed data, including the covariate x, and used this information to generated replacements for missing y that took its relation with x into account.
To see this, it is worth looking at a comparison of the observed and the imputed data.
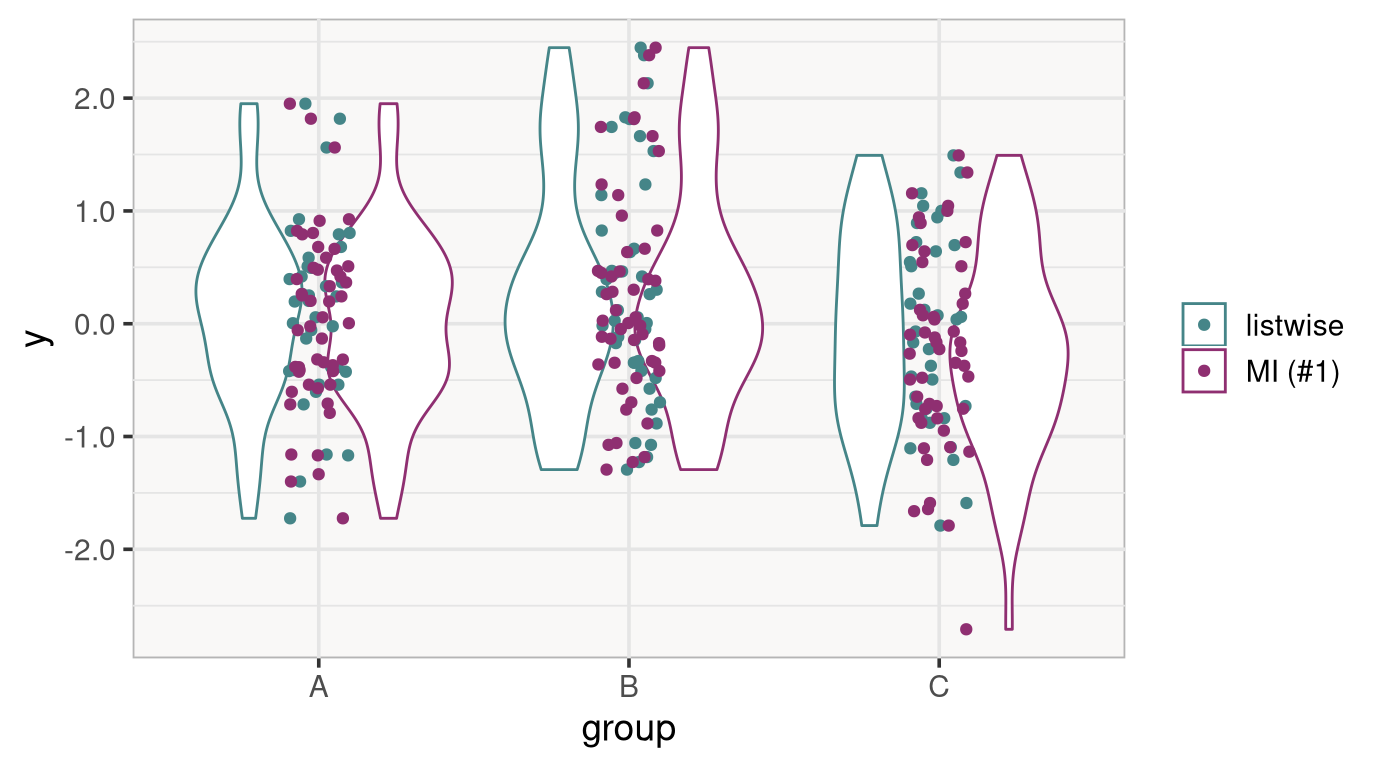
The difference is not extreme, but it is easy to see that the imputed data tend to have more mass at the lower end of the distribution of y (especially in groups A and C).
This is again a result of how the data were simulated: Lower y values, through their relation with x, are missing more often, which is accounted for using MI.
Conversely, using listwise deletion placed the group means more closely together than they should be, and this affected the results in the ANOVA.
Example 2: mixed/within-subjects ANOVA
In a within-subjects design, the design factor varies within (not between) persons, and we obtain multiple, repeated measurements for each condition (mixed designs include both). Fortunately, the procedure for the treatment and missing data and the analysis remains mostly the same.
Although within-subjects designs are analyzed most often with the repeated-measures ANOVA, mixed-effects models have become a popular alternative. Here, I will choose the latter because mixed-effects models make it straightforward to pool ANOVA-like hypotheses in within-subjects designs.
To fit the mixed-effects model, we will use the lmer() function from the package lme4.
library(lme4)I simulated a second data set similar to the one above, with \(n\) = 20 persons, a within-subject factor with three conditions, and five repeated measurements for each condition (CSV, Rdata).
| id | cond | y | x | |
|---|---|---|---|---|
| 1 | 1 | A | 1.318 | -0.235 |
| 2 | 1 | B | -0.085 | -0.235 |
| 3 | 1 | C | -0.040 | -0.235 |
| 4 | 1 | A | 0.680 | -0.235 |
| 5 | 1 | B | 0.344 | -0.235 |
The variables mean the following:
id: the subject identifiercond: the grouping variable (within subjects)y: the repeated measurements for the outcome variable (with 16% missing data)x: a subject-specific covariate
Like above, persons with lower values in x had a higher chance of missing data in y. Notice that cond varies within subjects, making the repeated measures for each condition “nested” within subjects.
Multiple imputation
To properly accommodate the “nested” structure of the repeated measurements, the imputation model can no longer be a simple regression. Instead, it needs to accommodate this structure by also employing a mixed-effects model. Specifying this model is easiest by first initializing the imputation model with the default values.
# run MI (for starting solution)
ini <- mice(data = dat, maxit = 0)Then we define the subject identifier for the imputation model (id) and change the imputation method to a use a mixed-effects model ("2l.pan").
Running MI is then the same as before.
# define the 'subject' identifier (code as '-2' in predictor matrix)
pred <- ini$pred
pred["y", "id"] <- -2
# run MI
imp2 <- mice(data = dat, pred = pred, method = "2l.pan", m = 100)
summary(imp2)The list of imputed data sets is generated as above.
# create a list of completed data sets
implist2 <- mids2mitml.list(imp2)The ANOVA model is then fit using lmer().
Notice that this model contains an additional term, (1|id), which specifies a random effect for each subject.
This effect captures unsystematic differences between subjects, thus accounting for the nested structure of the repeated-measures data.
# fit the ANOVA model
fit2 <- with(implist2, lmer(y ~ 1 + cond + (1|id)))
testEstimates(fit2, var.comp = TRUE)#
# Call:
#
# testEstimates(model = fit2, var.comp = TRUE)
#
# Final parameter estimates and inferences obtained from 100 imputed data sets.
#
# Estimate Std.Error t.value df P(>|t|) RIV FMI
# (Intercept) 0.069 0.161 0.428 42782.449 0.668 0.051 0.048
# condB 0.217 0.107 2.026 2738.362 0.043 0.235 0.191
# condC -0.061 0.107 -0.569 2786.073 0.569 0.232 0.189
#
# Estimate
# Intercept~~Intercept|id 0.399
# Residual~~Residual 0.466
# ICC|id 0.461
#
# Unadjusted hypothesis test as appropriate in larger samples.The output is similar to before, with the regression coefficients denoting the differences between the conditions. In addition, the output includes the variance of the random effect that denotes the unsystematic differences between subjects.
Testing the null hypothesis of the ANOVA again requires the specification of a reduced model that does not contain the parameters to be tested (i.e., those pertaining to cond).
# pool the parameter estimates
fit2.reduced <- with(implist2, lmer(y ~ 1 + (1|id)))
testModels(fit2, fit2.reduced, method = "D1")#
# Call:
#
# testModels(model = fit2, null.model = fit2.reduced, method = "D1")
#
# Model comparison calculated from 100 imputed data sets.
# Combination method: D1
#
# F.value df1 df2 P(>F) RIV
# 3.738 2 5512.706 0.024 0.229
#
# Unadjusted hypothesis test as appropriate in larger samples.As in the first example, the three conditions are significantly different from one another.
These two examples were obviously very simple. However, the same general procedure can be used for more complex ANOVA models, including models with two or more factors, interaction effects, or for mixed designs with both between- and within-subject factors.
Update: Post-hoc contrasts
In the one-way ANOVA, testing the (omnibus) null hypothesis is often only one part of the journey. Once an omnibus test yielded a positive result, indicating that the means differ between groups, we often want to know which groups differ by how much. This can be done by testing post-hoc contrasts.
For example, suppose we have conducted the between-subjects ANOVA with three groups—labeled A, B, and C—in Example 1. The omnibus \(F\)-test is significant, but which groups differ the most from each other?
In the following, we will look at two examples for testing contrasts between groups on the basis of the imputed data obtained from MI.
Pairwise contrasts
The easiest option for testing pairwise contrasts between groups is to use one of the R packages that exist for this purpose.
For this example, we will use the multcomp package.3
library(multcomp)In the multcomp package, the glht() function is used to test post-hoc contrasts.
Here, we will use Tukey’s method to test pairwise contrasts between the groups (i.e., B vs. A, C vs. A, and C vs. B)
Based on the imputation conducted in Example 1 and ANOVA model fitted to the 100 imputed data sets (fit1), we can use the lapply function to apply glht() to all of the imputed data sets.
This will perform pairwise contrasts for all data sets simultaneously.
# perform pairwise comparisons (Tukey)
fit1.pairwise <- lapply(fit1, glht, linfct = mcp(group = "Tukey"))Next, the parameter estimates must be pooled across the imputed data sets.
The object created by lapply, however, is only a simple list and needs to be converted into a mitml.result object.
Then, we can pool the results as usual.
# convert to "mitml.result" and pool the parameter estimates
fit1.pairwise <- as.mitml.result(fit1.pairwise)
testEstimates(fit1.pairwise)#
# Call:
#
# testEstimates(model = fit1.pairwise)
#
# Final parameter estimates and inferences obtained from 100 imputed data sets.
#
# Estimate Std.Error t.value df P(>|t|) RIV FMI
# B - A 0.204 0.200 1.022 4816.135 0.307 0.167 0.144
# C - A -0.328 0.204 -1.611 3274.908 0.107 0.210 0.174
# C - B -0.532 0.202 -2.639 3968.657 0.008 0.188 0.158
#
# Unadjusted hypothesis test as appropriate in larger samples.In this example, it seems that the means differ primarily between groups B and C, whereas those between the groups B and A and C and A, respectively, are somewhat smaller and not statistically significant.
Other contrasts
The same method can be used to test more specific hypotheses.
For example, we may be interested in how the mean in one group (say B) compares with the means in the other two groups (say A and C).
Such a specific hypothesis can be tested with glht() by providing a symbolic representation of the contrast.
# specify contrast comparing group B with groups A and C
fit1.B.vs.AC <- lapply(fit1, glht, linfct = mcp(group = "B - (A + C)/2 = 0"))The resulting object is then converted to the mitml.result format, and the results are pooled as before.
# convert to "mitml.result" and pool the parameter estimates
fit1.B.vs.AC <- as.mitml.result(fit1.B.vs.AC)
testEstimates(fit1.B.vs.AC)#
# Call:
#
# testEstimates(model = fit1.B.vs.AC)
#
# Final parameter estimates and inferences obtained from 100 imputed data sets.
#
# Estimate Std.Error t.value df P(>|t|) RIV FMI
# B - (A + C)/2 0.368 0.173 2.128 4860.807 0.033 0.166 0.143
#
# Unadjusted hypothesis test as appropriate in larger samples.This contrast, too, appears to be statistically significant (although just barely so).
Note on multiple comparisons
In many cases, it makes sense to correct for multiple comparisons when testing post-hoc contrasts, for example by applying corrections to the \(p\)-values obtained from multiple tests or by adjusting the \(\alpha\) level with which these are compared.
Unfortunately, software solutions that provide these corrections for multiply imputed data are in short supply. For this reason, these corrections need to be applied “by hand.” For example, to apply a Bonferroni correction, we would simply follow the steps outlined above but adjust the \(\alpha\) level with which we compare the \(p\)-values in the pooled results obtained from MI.
Procedures other than MI
Imputation is not the only method that can deal with missing data, and other methods like maximum-likelihood estimation (ML) have also been recommended (Schafer & Graham, 2002). Using ML, cases contribute to the estimation of the model only to the extent to which they have data, and its results are often equally trustworthy as those under MI.
However, in the ANOVA, this should be taken with a grain of salt.
For missing data in the outcome variable y, using ML simply means that the model is estimated using only the cases with observed y (i.e., listwise deletion), which can lead to distorted parameter estimates if other variables are related to the chance of observing y (see Example 1).
In order to account for this, ML requires including these extra variables in the analysis model, which changes the meaning of the parameters (i.e., the ANOVA becomes ANCOVA, though the estimates for it would be unbiased!).
One key advantage of MI is that the treatment of missing data is independent of the analysis. Variables relevant for the treatment of missing data can be included in the imputation model without altering the analysis model.
Further reading
To read more about ANOVA models and the treatment of missing data therein, you can check the following resources:
- Maxwell, Delaney, and Kelley (2018) give a great introduction into the design and analysis of experimental data with the ANOVA and mixed-effects models
- van Ginkel and Kroonenberg (2014) provide a detailed discussion of missing data and MI in the ANOVA with examples, syntax files, and a macro for SPSS
- Grund, Lüdtke, and Robitzsch (2016) provide a comparison of different methods for testing hypotheses in the ANOVA under MI
- Liu and Enders (2017) provide a similar comparison in the context of regression analyses
References
- Cohen, J. (1988). Statistical power analysis for the behavioral sciences (2nd ed.). Hillsdale, NJ: Erlbaum.
- Grund, S., Lüdtke, O., & Robitzsch, A. (2016). Pooling ANOVA results from multiply imputed datasets: A simulation study. Methodology, 12, 75–88. doi:10.1027/1614-2241/a000111
- Liu, Y., & Enders, C. K. (2017). Evaluation of multi-parameter test statistics for multiple imputation. Multivariate Behavioral Research, 52, 371–390. doi:10.1080/00273171.2017.1298432
- Maxwell, S. E., Delaney, H. D., & Kelley, K. (2018). Designing experiments and analyzing data: A model comparison perspective (3rd ed.). Mahwah, NJ: Erlbaum.
- Schafer, J. L., & Graham, J. W. (2002). Missing data: Our view of the state of the art. Psychological Methods, 7, 147–177. doi:10.1037//1082-989X.7.2.147
- van Ginkel, J. R., & Kroonenberg, P. M. (2014). Analysis of variance of multiply imputed data. Multivariate Behavioral Research, 49, 78–91. doi:10.1080/00273171.2013.855890
The hypothesis test in ANOVA is a Wald test that simultaneously tests all the differences between groups against zero. In this example, these differences are represented by the regression coefficients for
groupBandgroupC.This can easily be verified by calculating the Wald test by hand:
# estimates and covariance matrix b <- coef(fit0)[-1] V <- vcov(fit0)[-1,-1] # Wald-test F <- b %*% solve(V) %*% b / 2 # F statistic pf(F, 2, 116, lower.tail = FALSE) # p value# [,1] # [1,] 0.26The resulting \(F\) and \(p\) value are exactly the same as in the output above.↩︎
Technically, a reduced model is not necessary (only convenient). The Wald test can be formulated equivalently with a linear constraint on the parameters of the full model (i.e., setting them to zero).
Under MI, this can be done, too, with the
testConstraints()function:# define and test parameter constraints con <- c("groupB", "groupC") testConstraints(fit1, constraints = con, method = "D1")# # Call: # # testConstraints(model = fit1, constraints = con, method = "D1") # # Hypothesis test calculated from 100 imputed data sets. The following # constraints were specified: # # Estimate Std. Error # groupB: 0.204 0.202 # groupC: -0.328 0.202 # # Combination method: D1 # # F.value df1 df2 P(>F) RIV # 3.543 2 7588.022 0.029 0.188 # # Unadjusted hypothesis test as appropriate in larger samples.The results of this are identical to those of
testModels().↩︎Post-hoc contrasts can also be tested with the
mitmlpackage by using the functiontestConstraints(), without the need for additional packages.For example, the same results as those for the pairwise contrasts above can also be obtained as follows:
# contrast between B and A testConstraints(fit1, constraints = "groupB")# # Call: # # testConstraints(model = fit1, constraints = "groupB") # # Hypothesis test calculated from 100 imputed data sets. The following # constraints were specified: # # Estimate Std. Error # groupB: 0.204 0.200 # # Combination method: D1 # # F.value df1 df2 P(>F) RIV # 1.044 1 4466.970 0.307 0.167 # # Unadjusted hypothesis test as appropriate in larger samples.# contrast between C and A testConstraints(fit1, constraints = "groupC")# # Call: # # testConstraints(model = fit1, constraints = "groupC") # # Hypothesis test calculated from 100 imputed data sets. The following # constraints were specified: # # Estimate Std. Error # groupC: -0.328 0.204 # # Combination method: D1 # # F.value df1 df2 P(>F) RIV # 2.594 1 3042.567 0.107 0.210 # # Unadjusted hypothesis test as appropriate in larger samples.# contrast between C and B testConstraints(fit1, constraints = "groupC - groupB")
↩︎# # Call: # # testConstraints(model = fit1, constraints = "groupC - groupB") # # Hypothesis test calculated from 100 imputed data sets. The following # constraints were specified: # # Estimate Std. Error # groupC - groupB: -0.532 0.202 # # Combination method: D1 # # F.value df1 df2 P(>F) RIV # 6.966 1 3683.841 0.008 0.188 # # Unadjusted hypothesis test as appropriate in larger samples.